Selected Publications
Complete publication list on Google Scholar
Braga, M. P., Janz, N., Nylin, S., Ronquist, F., & Landis, M. J. (2021). Phylogenetic reconstruction of ancestral ecological networks through time for pierid butterflies and their host plants. bioRxiv.
Landis, M. J., Eaton, D. A. R., Clement, W. L., Park, B., Spriggs, E. L., Sweeney, P. W., Edwards, E. J., & Donoghue, M. J. (2021). Joint phylogenetic estimation of geographic movements and biome shifts during the global diversification of Viburnum. Systematic Biology 70: 67-85.
Landis, M., Edwards, E. J., & Donoghue, M. J. (2021). Modeling phylogenetic biome shifts on a planet with a past. Systematic Biology, 70: 86-107.
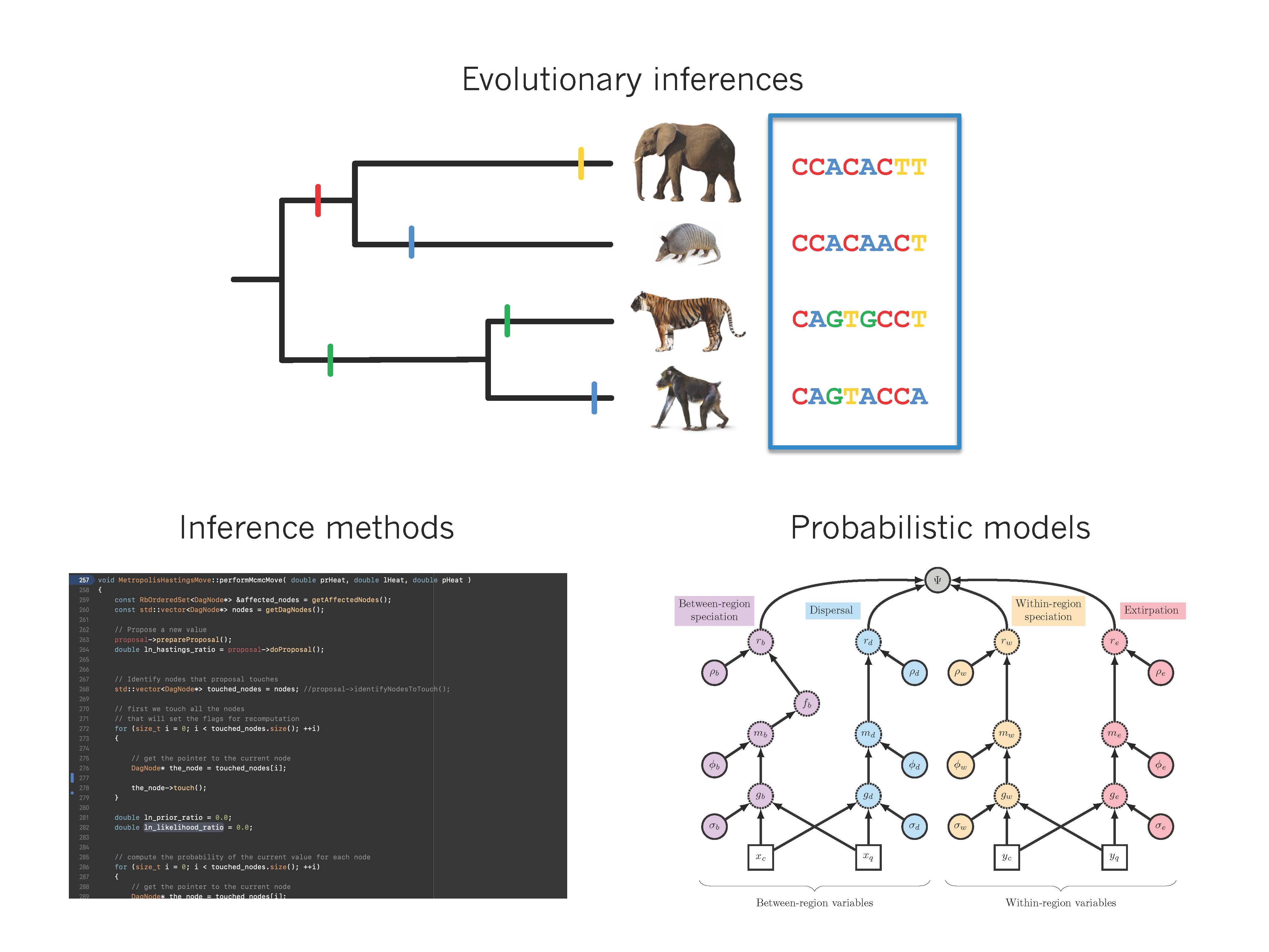
Landis, M. J. (2020). Biogeographic Dating of Phylogenetic Divergence Times Using Priors and Processes. In The Molecular Evolutionary Clock (pp. 135-155). Springer, Cham.
Braga, M. P., Landis, M. J., Nylin, S., Janz, N., & Ronquist, F. (2020). Bayesian Inference of Ancestral Host–Parasite Interactions under a Phylogenetic Model of Host Repertoire Evolution. Systematic Biology 69: 1149-1162.
Kim, A. S., Zimmerman, O., Fox, J. M., Nelson, C. A., Basore, K., Zhang, R., Durnell, L., Desari, C., Deem, S. L., Oppenheimer, J., Shapiro, B., Wang, T., Cherry, S., Coyne, C. B., Handley, S. A., Landis, M. J., Fremont, D. H., & Diamond, M. S. (2020). An evolutionary insertion in the Mxra8 receptor-binding site confers resistance to alphavirus infection and pathogenesis. Cell Host & Microbe 27: 428-440.
Quintero, I., & Landis, M. J. (2020). Interdependent phenotypic and biogeographic evolution driven by biotic interactions. Systematic Biology 69: 739–755.
Landis, M. J., Freyman, W. A., & Baldwin, B. G. (2018). Retracing the Hawaiian silversword radiation despite phylogenetic, biogeographic, and paleogeographic uncertainty. Evolution, 72: 2343-2359.
Landis, M. J., & Schraiber, J. G. (2017). Pulsed evolution shaped modern vertebrate body sizes. Proceedings of the National Academy of Sciences 114: 13224-13229.
Höhna, S., Landis, M. J., Heath, T. A., Boussau, B., Lartillot, N., Moore, B. R., Huelsenbeck J. P., & Ronquist, F. (2016). RevBayes: Bayesian phylogenetic inference using graphical models and an interactive model-specification language. Systematic Biology 65: 726-736.
Schraiber, J. G., & Landis, M. J. (2015). Sensitivity of quantitative traits to mutational effects and number of loci. Theoretical Population Biology: 102, 85-93.
Landis, M. J., & Bedford, T. (2014). Phylowood: interactive web-based animations of biogeographic and phylogeographic histories. Bioinformatics 30: 123-124.
Landis, M. J., Matzke, N. J., Moore, B. R., & Huelsenbeck, J. P. (2013). Bayesian analysis of biogeography when the number of areas is large. Systematic Biology, 62: 789-804.
Landis, M. J., Schraiber, J. G., & Liang, M. (2013). Phylogenetic analysis using Lévy processes: finding jumps in the evolution of continuous traits. Systematic Biology 62: 193-204.